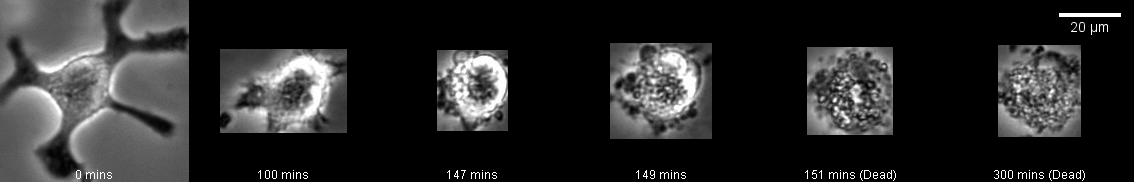
Cell Death Detection
In order to identify when a macrophage dies, the Mask R-CNN model can be trained on images containing both alive and dead cells. In general here, the moment of cell death is taken as when the cell looses it’s structure and the membrane can no longer be clearly defined. (Cells often ball up and exhibit membrane blebbing before this occurs.)
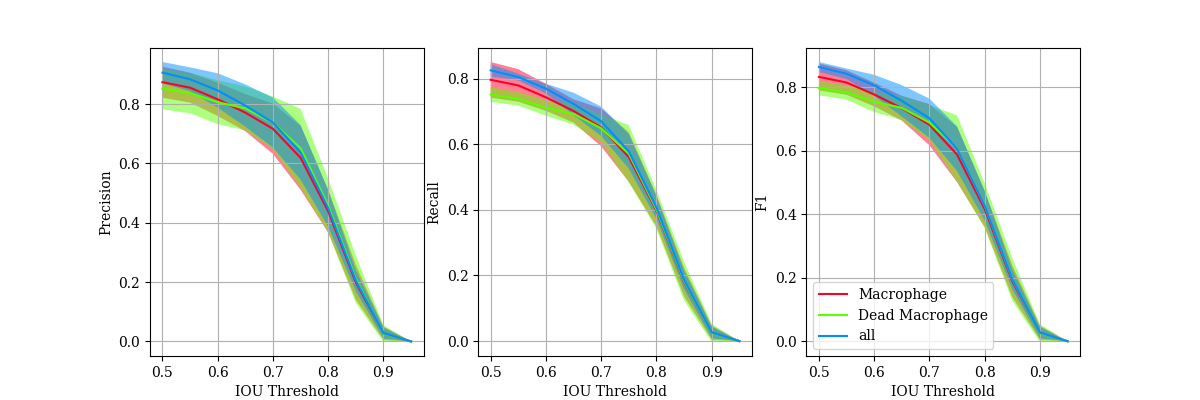
 KFold cross validation on the training images (taking one image out for validation per fold), suggested that this performed reasonably:
KFold cross validation on the training images (taking one image out for validation per fold), suggested that this performed reasonably:
 These classifications could then be used to identify the frame of cell death. It was assumed there would be some noise in the classifications, so alive cells were asigned a score 1, and dead cells 0. These scores were then smoothed for each cell over time (by taking a 5 frame rolling average) and the cell death was taken as the first frame for which the score dropped below 0.5 for at least 10 frames. However, as shown below, this did not accurately identify cell death. Average error was 56.5 frames.
These classifications could then be used to identify the frame of cell death. It was assumed there would be some noise in the classifications, so alive cells were asigned a score 1, and dead cells 0. These scores were then smoothed for each cell over time (by taking a 5 frame rolling average) and the cell death was taken as the first frame for which the score dropped below 0.5 for at least 10 frames. However, as shown below, this did not accurately identify cell death. Average error was 56.5 frames.
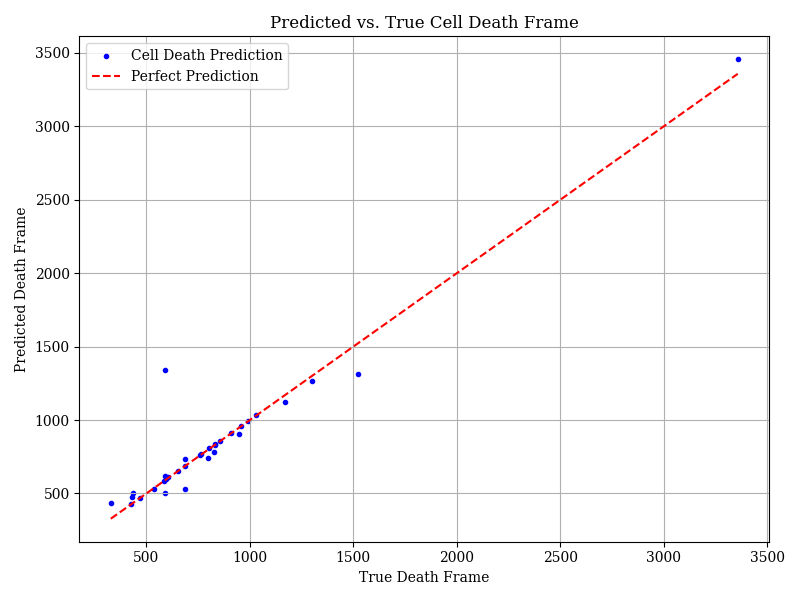 The training images contained both alive and dead cells. However, cell appearance changed significantly just before death. So to accurately identify the exact frame of death, training data of cells just before and just after death would be required.
For 50 cells, the exact frame of death was manually identified. Frames [1, 2, 3, 4, 5, 50, 100, 150, 200] before and after this were used to create a new fine tuning dataset. Images were cropped to the region of the cell to ensure all masks were correct. A training and validation set were crated with an 80/20 split. Below is the results of the original model, as well as the results after fine tuning:
The training images contained both alive and dead cells. However, cell appearance changed significantly just before death. So to accurately identify the exact frame of death, training data of cells just before and just after death would be required.
For 50 cells, the exact frame of death was manually identified. Frames [1, 2, 3, 4, 5, 50, 100, 150, 200] before and after this were used to create a new fine tuning dataset. Images were cropped to the region of the cell to ensure all masks were correct. A training and validation set were crated with an 80/20 split. Below is the results of the original model, as well as the results after fine tuning:
- Just the classificaiton head
- The whole ROI head
- The entire model
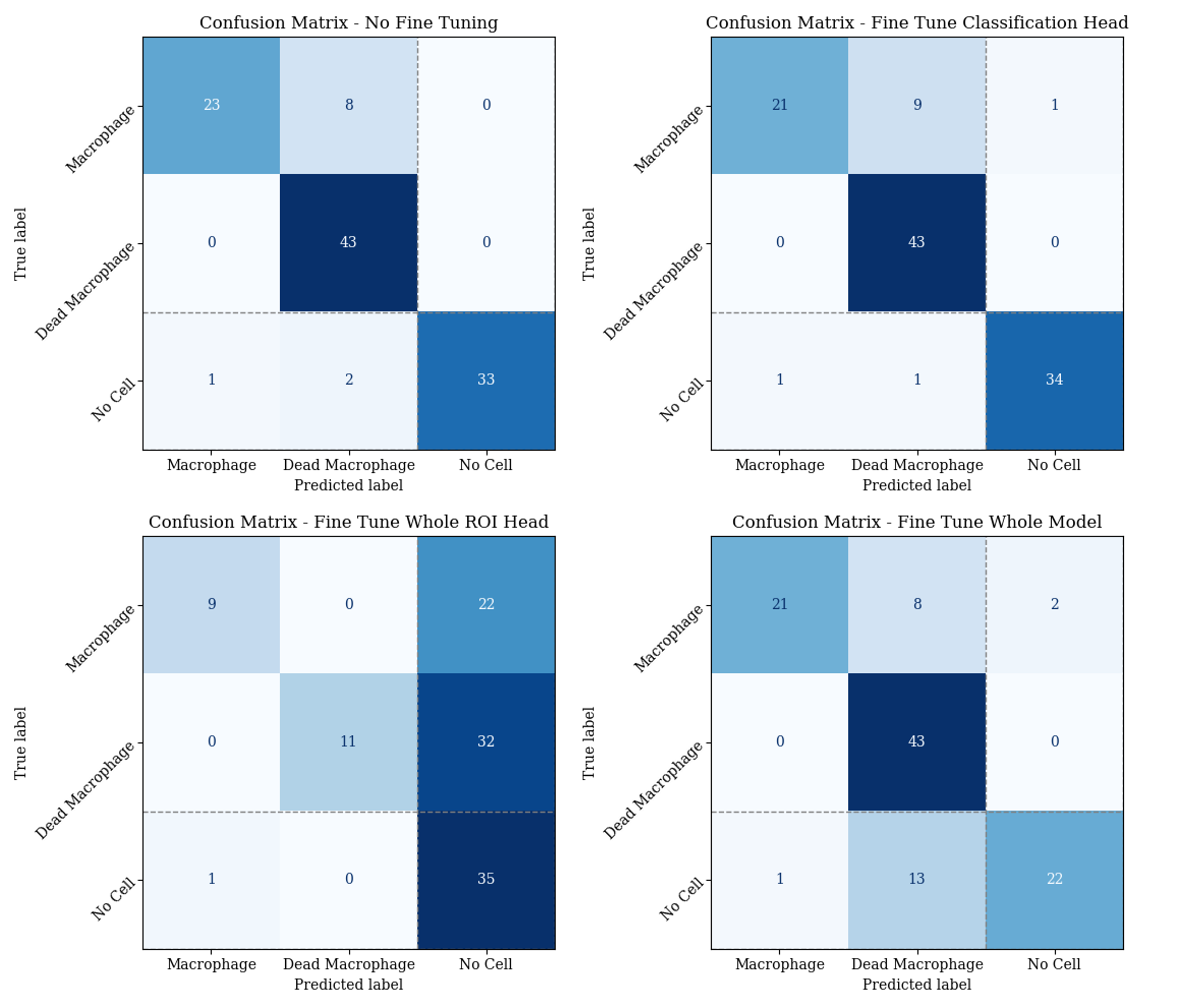 The resulting accuracies are either lower than or the same as when doing no fine tuning, more training data is probably needed.
More cells were added to the training set (total 200 cells). For each cell a random frame between 1 and 10 frames before cell death was selected, a frame 100 frames before cell death and a random frame between1 and 10 frames after cell death. False psitive segmentations and incorrectly merged cells (undersegmentation) were also corrected and added to the dataset.
The resulting accuracies are either lower than or the same as when doing no fine tuning, more training data is probably needed.
More cells were added to the training set (total 200 cells). For each cell a random frame between 1 and 10 frames before cell death was selected, a frame 100 frames before cell death and a random frame between1 and 10 frames after cell death. False psitive segmentations and incorrectly merged cells (undersegmentation) were also corrected and added to the dataset.