Feature Extraction
PhagoPred.feature_extraction.extract_features
Features describing the morphology and dynamics of each cell are then extracted. These are classed as either primary or secondary features depending on the information required to compute them. All feature classes inherit from PhagoPred.feature_extraction.features.BaseFeature() class and must have a compute() method.
Primary Features
def compute(self, mask: torch.tensor, image: torch.tensor) -> np.array:
These are calculated directly from the segmentation masks and raw images. They are computed for a batch of cells in each frame at a time to allow calculations to be performed on the GPU for faster speeds.
- Coords
- Perimeter
- Circularity
- Morphology Modes
- Gabor Scale
Secondary Features
def compute(self, phase_xr: xr.Dataset, epi_xr: xr.Dataset) -> np.array:
These are calculated from other features. They take the datasets of all other features as their arguments, stored as xarray Datasets backed with Dask (allowing parallelised computation with automatic batching).
- Speed
- Displacement
- Surrounding cell density
Examples
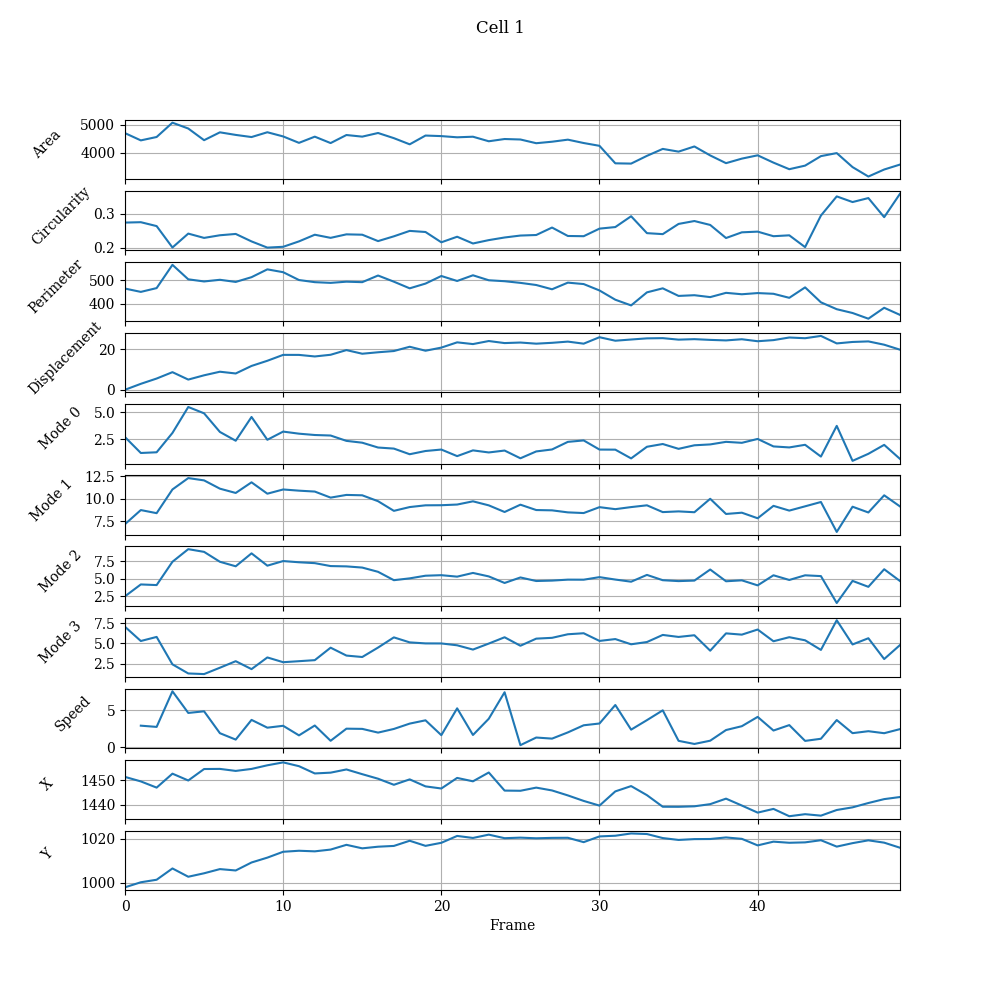
Individual Cell
 |
 |
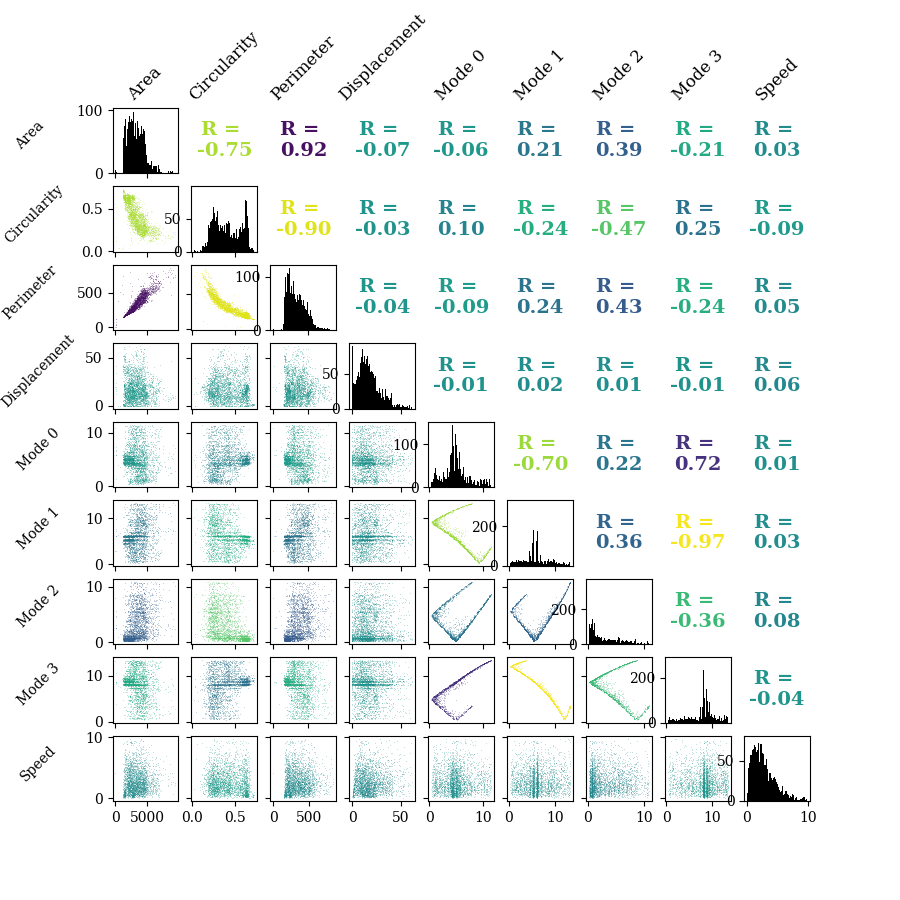
Correlation Plot
PhagoPred.display.plots.plot_feature_correlations()
Correlation plot of a subset of the features. R gives the correlation coefficient between each pair of features and histograms of each feature are shown on the diagonal.